Current projects involve the extraction, modification, and functionalization of various plant proteins following novel approaches, while also investigating flavor and nutritional quality. Projects aim at relating structural characteristics to functional properties, bioactivity, and allergenicity. The projects target the characterization and modification of novel plant protein ingredients and protein in waste streams for utilization in unique and high-end applications.
Click the links below to jump to the project description.
PPIC Funded Projects:
- Plant Protein Blending: Inducing Molecular Interactions to Enhance Texturization (Pea, Chickpea, Oat)
- Impact of Pea Storage Protein Fractions and Their Ratio on Functionality and Nutritional Quality
- Enhancing Pennycress Oilseeds as a New Protein Source by Improving Flavor and Protein Extractability
- Flavor Reactions with Plant Proteins (Pea)
- Impact of Cold Plasma Treatment on Pea Protein Structural and Functional Characteristics
- An Interdisciplinary Strategy for Improving Hemp Protein as a Food Ingredient through Plant Breeding and Processing
Externally Funded Ongoing Protein Research Projects:
- Characterizing and Texturizing Proteins from Pulses to Form Fibers with Textures that Mimic Chicken
- Legumes of the Future: Developing Methodologies and Germplasm to Enhance the Functionality and Nutritional Quality of Pea Protein
- Optimization of Extraction of Camelina (Camelina sativa) Protein and Screening for Varietal Differences
- Impact of Selective Breeding, Extraction Conditions, and Modification Technologies on Pennycress Protein Structural, Functional, and Nutritional Characteristics
- Protein profiling and characterization of diverse oat lines
- Extraction and Modification of Pea Protein for Improved Functionality, Flavor, and Nutrition
- Functionalization of Plant Proteins
- Protein-Flavor Interactions in Food Matrices
Externally Funded Completed Protein Research Projects:
- Enzymatic modification of pea protein isolate for improved functionality
- Improving the Functionality of Low Protein Streams for High Value Applications
- Extraction, Modification, and Structural/Functional Characterization of Camelina Protein
- The Effect of Limited Enzymatic Hydrolysis and Maillard-Induced Glycation on Soy Protein Immunoreactivity and Functionality
- Proteomics Approach to Characterize Intermediate Wheatgrass Proteins
- Identification of key volatile compounds contributing to pennycress aroma
PPIC Funded Research Projects
Plant Protein Blending: Inducing Molecular Interactions to Enhance Texturization
Co-PIs: Daniel Gallaher, Job Ubbink, Zata Vickers
Researcher: Brittany Kralik
Impact of Pea Storage Protein Fractions and Their Ratio on Functionality and Nutritional Quality
Principal investigator: B. Pam Ismail
Co-PIs: Daniel Gallaher, Robert Stupar
Researcher: Holly Husband
The overall goal of this project is to determine the impact of varying the proportion of pea storage proteins on functional and nutritional properties for enhanced breeding efforts. Storage pea protein components, namely legumin, vicilin, and convicilin, as well as the albumin fraction, will be separated and purified. The isolated fractions will be reconstituted into protein isolates in different ratios (from 0 to 1 for each fraction). Separated fractions as well as reconstituted protein isolates will be subjected to structural characterization, including protein denaturation, protein profiling, surface charge, surface hydrophobicity, and protein secondary structure. Additionally, the functional properties (solubility, emulsification, foaming, and gelation) of the fractions and isolates will be determined. Commercial PPI, lab produced PPI, and commercial SPI will be used as references. The in vitro PDCAAS will be determined for each of the fractions and isolates, using a newly available commercial kit. Data generated will aid in determining the association between different pea protein components and functional properties, as well as identifying the optimal protein profile. Understanding this association is critical when screening for pea protein cultivars that could potentially have superior functionality and nutritional quality, which could be used as parental cultivars in breeding programs aimed at improving the functional aspects of pea protein and not just yield.
Update:
Since beginning this project, the conditions of alkaline protein extraction have been optimized to produce pea protein isolate from pea flour with ideal purity and yield. This optimization is necessary to supply the pea protein isolate needed as a reference sample when analyzing the molecular and structural properties, functionality, and nutritional quality of fractionated pea protein isolate. The next phase of the project is in progress and is focused on the selection of methodology for pea protein fractionation. Various methods of fractionation including alkaline separation, size-exclusion chromatography, and membrane filtration will be evaluated to select the most efficient fractionation method. Work is currently being done on alkaline separation with reference to a published method for soy protein fractionation. If similar fractionation results are observed using pea, this method may be chosen as the final fractionation method for the project. (03/2021)
Enhancing Pennycress Oilseeds as a New Protein Source by Improving Flavor and Protein Extractability
Principal investigator: M. David Marks
Co-PIs: B. Pam Ismail, Gary Reineccius, Adrian Hegeman, Ratan Chopra, Krishan Mohan Rai
Researcher: Ratan Chopra & Devon McDonald
Pennycress is being domesticated as a new eco-friendly winter/spring oilseed. Ongoing work has led to the development of pennycress varieties that produce canola-like oil. This oil can be used for human nutrition or can be used for the creation of biofuels and products. Additional work has identified varieties that have reduced glucosinolates and reduced condensed tannins. We propose to make further enhancements in pennycress that will allow it to become a new source of proteins for human use. Current varieties of pennycress contain chemicals such as phytic acid, sinapine, and dimethyl methoxypyrazine that adversely affect the flavor and function of plant proteins. The separation of these compounds from plant proteins requires extra extraction procedures that can alter protein functionality and increase processing costs. We have created a large pool of mutant lines that have been characterized by whole genome sequencing, and within this pool we have identified lines that are predicted to have reductions in the undesirable compounds. The proposed changes to pennycress seed chemistry will greatly enhance the value and utility of pennycress seed proteins. We will present these new pennycress varieties to the food science members of our team for testing.
Update:
The winter annual plant species pennycress is being domesticated as a new cash/cover crop that can protect farmlands between the harvest and establishment of traditional summer crops and provide farmers with a new source of income. To enhance the value of pennycress seed meal efforts are being made to improve pennycress meal flavor. Candidate pennycress varieties have been identified that may reduce the quantities of known compounds that adversely affect flavor. Currently, several of these varieties are being grown to produce seeds for flavor analyses. Sufficient seeds should be available this coming summer to begin testing these candidates.
The aroma profiles of pennycress and its extracts are being collected using the purge and trap method that offers solvent-free, highly reproducible results. The volatile compounds are collected from the seeds, defatted meal, and multiple protein extracts to show what the original profile is and how volatile flavor compounds change with different extraction techniques. The current line of pennycress still has considerable amounts of off-flavors, such as isothiocyanates, sulfides, and cyanides, that give it an undesirable mustard aroma. Current extraction protocols lower the influence of some of the undesirable compounds, but could possibly impart new or concentrated off-flavors that were not originally in the seed. Identifying all of these compounds that can be minimized to improve the flavor of pennycress products will be helpful for extraction and processing work in the future to turn pennycress into a desirable plant protein source. (03/2021)
Flavor Reactions with Plant Proteins
Principal investigator: Gary Reineccius
Co-PIs: B. Pam Ismail, Daniel Gallaher
Researcher: Igor Shepelevs
The overall goal of this work is to develop analytical methodology to measure the covalent bonding that occurs between flavor compounds and the very complex and diverse plant proteins (this project will focus on pea protein). There are two competing approaches to achieve our goals – radioisotope labelling and mass spectrometry (MS). The MS approach begins with an enzyme digestion and then uses tandem mass spectrometry to identify the sites of post translational modifications that take place on the various proteins after reacting with a flavor molecule. A targeted m/z approach on a triple quad LC/MS will be used for quantitation. The alternative and potentially complimentary approach is to use radioactively labeled flavor compounds to measure covalent bond formation. A radio labelled flavor compound will be reacted with pea protein isolate (PPI), the unreacted isotopes separated from the protein, and the radioactivity will be measured with a scintillation counter. The MS method has the advantage of providing an understanding of where flavor compounds react on each protein even in a mixture of proteins. This knowledge can provide information that will help us design ways to mitigate these reactions. The radioisotope approach has the advantage of being a rapid, highly quantitative measure of the degree of bonding with whatever proteins are present in the sample.
Update:
The objective of this project is to develop a method to accurately measure the covalent reactions that occur between flavor molecules and plant proteins. These reactions result minimally in undesirable changes in flavor character and in many cases, in flavor loss that can bring about end of shelf-life. We have a LC-MS method that is well suited to measure covalent reactions with small proteins but not large proteins common to plants. Thus, we have been working on a radioisotope method whereby we can get good measurements of flavor – protein reactions with larger (plant) proteins. Our method of choice is to use C14 labelled flavor molecules to monitor this reaction.
In principle, one would simply add a C14 labelled flavor molecule to a protein, allow a reaction period, and then extract the unreacted flavor from the protein reaction system (commonly an aqueous system) using a non-polar solvent. Unfortunately, proteins bind flavors by two mechanisms: hydrophobic bonding and covalent bonding. The hydrophobic bonding can be sufficiently strong to make extraction uncertain. Also, one cannot solvent extract most protein-containing systems since they typically serve as good emulsifiers which creates a stable solvent: water emulsion that cannot be separated easily. After much effort, we have been able to work with low protein concentrations, salt addition and pH adjustments to keep the proteins from forming stable emulsions. We have just received two radiolabeled flavor molecules: one to serve as a control that will bind via hydrogen bonding and a second that bonds primarily by covalent reaction. We will be testing the extraction method the week of April 5. (03/2021)
Impact of Cold Plasma Treatment on Pea Protein Structural and Functional Characteristics
Principal investigator: B. Pam Ismail
Co-PIs: Peter Bruggeman, George Annor, Chi Chen
Researcher: Fan Bu
While pea protein is gaining traction, functionality limitations are hindering its market growth. Improving pea protein functionality will enable its successful utilization in various food products. There are several reports on pea protein functionality and applications, but much is still not known about the effect of different processing and modifications on the structural and associated functional changes in pea protein. Cold plasma, a non-thermal processing technique, is being explored as a novel means for protein functionalization. Cold plasma technology involves the exposure of plasma, a partially ionized gas, to proteins. The reactive species generated by cold plasma can induce several chemical reactions including oxidation, bond cleavage, and/or polymerization. The overall goal of this project is to evaluate the effect of various cold plasma treatments on pea protein structural and functional properties. Reactive species and changes due to potential chemical reactions will be monitored. Specifically, changes in the protein tertiary, secondary, and primary structure will be evaluated, and chemical reactions will be elucidated. The impact of structural change on protein functionality will be determined. This research will provide for the first time a controlled evaluation of the impact of cold plasma on pea protein structural and functional characteristics. Cold plasma treatment may lead to the production of a viable pea protein ingredient with functional properties that are comparable or better than those of traditional protein ingredients. The information gathered from this short-term project, as well as the joined interdisciplinary effort will definitely lead to designing future long-term studies with a targeted outcome.
Update:
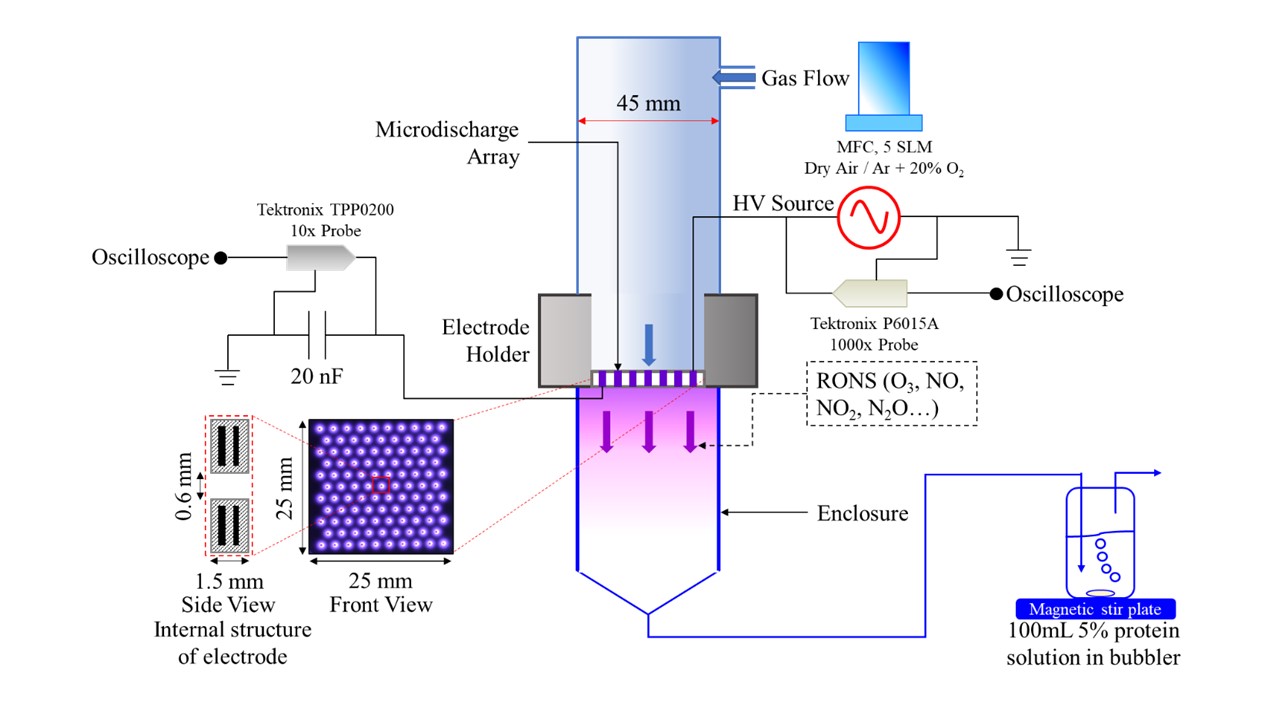
Pea (Pisum sativum) protein is gaining more interest as a potential replacement of high-allergenicity soy, whey, and egg protein in food applications. However, limitations in functionality limit the use of pea protein isolate in food products. Cold atmospheric plasma (CAP), a non-thermal processing and anti-microbial technology is a novel method that can be used to change the primary, secondary, and tertiary structure of protein. OH radical, H2O2, O3, and reactive nitrogen species (RNS) are major components in plasma. Plasma generated with different gas mixtures and by different configurations results in different profile of reactive species, leading to various changes in protein structure and consequently protein functionality. Therefore, the objectives of this project were to 1) investigate the impact of OH radical, H2O2, O3, and RNS on the structure and functionality of pea protein isolate, and 2) investigate the impact of atmospheric pressure plasma jet (APPJ), dielectric barrier discharge (DBD), and nanosecond pulsed discharge (ns-pulsed) on the structure and functionality of pea protein isolate. Pronounced structural and functional effects were observed when PPI was treated with plasma reactive species at pH 2 compared to pH 7. All plasma species induced the formation of disulfide-linked soluble aggregates with molecular weights around 1200 kDa, as demonstrated by SE-HPLC and SDS-PAGE. Protein denaturation was observed after treatment with all plasma species. However, a significant increase in β-sheet content and surface hydrophobicity was only induced by treatment with O3 and OH, which resulted in the greatest enhancement in gelation and emulsification properties of PPI. Additionally, we found that the gel strength and emulsification capacity of PPI were significantly improved after all plasma treatments whereas the solubility of PPI was decreased significantly. Denaturation enthalpy of all APPJ-, DBD-, and ns-pulsed- treated PPI were significantly lower than that of non-treated PPI, indicating that plasma triggered the unfolding of protein. The significant incremental increase in soluble aggregates content after plasma treatment was revealed by SE-HPLC. Overall, DBD- 30min was the optimal plasma modification condition for improving gel strength and emulsification capacity of pea protein isolates. Soluble pea protein aggregates obtained as a result of plasma treatment can potentially result in enhanced texturization potential for the formation of fibrous meat-like structures. Future work will focus on correlating findings to texturizing properties. (03/2021)
An Interdisciplinary Strategy for Improving Hemp Protein as a Food Ingredient through Plant Breeding and Processing
Principal investigator: Tom Michaels
Co-PIs: B. Pam Ismail, James House
Researcher: Laura Echkardt
Hemp seed protein benefits human diets through its high digestibility, potential free radical-scavenging activity, and hypoallergenicity. However, it is considered an incomplete protein due to limiting amino acids lysine, leucine, and tryptophan. In food preparations, hemp protein exhibits poor solubility, relative to soybean protein. Genetic variance among current industrial hemp genotypes for protein nutritional quality and structural/functional properties for food applications is unknown. Therefore, the potential for breeding new industrial hemp varieties with improved protein characteristics is unexplored. The objectives of this project are to develop hemp seed protein extraction protocols, determine protein quantity, nutritional quality and structural/functional properties for food applications, and identify differences among industrial hemp genotypes and breeding lines for these characteristics. Once these protocols have been optimized and genetic differences characterized, our team will develop a pragmatic, interdisciplinary strategy for improving hemp protein as a food ingredient that integrates plant breeding and optimization of processing protocols. Our team will then pursue this strategy in subsequent research projects.
Update:
The conditions for optimized protein extraction using alkaline solubilization/isoelectric precipitation and salt solubilization coupled with membrane filtration have been determined. Extraction at pH 11 resulted in higher protein purity (88% protein) and yield (87% yield) compared to extraction at pH 10. The protein purity and yield are quite high, which indicates industrial feasibility. Salt extraction using coupled with ultrafiltration similarly resulted in high and appreciable protein purity and yield. Both isolates have a bland off white color, which is also desirable. These protein extraction methods have been used to produce two larger batches of hemp protein isolate (HPI) for characterizing structural and functional attributes. Soy protein isolate (SPI) and pea protein isolate (PPI) produced by the Ismail lab has been procured along with commercially produced SPI and PPI as reference samples for the structure/function work. Structure/function work is currently underway. (03/2021)
Externally Funded Ongoing Protein Research Projects
Characterizing and Texturizing Proteins from Pulses to Form Fibers with Textures that Mimic Chicken
Funded by The Good Food Institute
Principal Investigators: Zata Vickers, Job Ubbink, & B. Pam Ismail
Researchers: Brigitta Yaputri & Kenzie Mcclintic
Pea protein has gained traction in the alternative meat sector in addition to soy and gluten. Chickpea is also gaining interest due to its low cost, high protein content (~20%), and good nutritional value. Though the application of pea protein in alternative meat products is increasing, pulse protein, in general, don’t have good texturization properties. Thus, there is a need to develop functionalization techniques. In addition, protein isolation method plays a fundamental role in dictating the structural and functionality properties of the protein. Production of pea protein isolates (PPI) and chickpea protein isolates (ChPI) are commonly done via pH-based extraction, but no data have been collected from salt extraction process coupled with ultrafiltration/ diafiltration.
Therefore, the first objective of this research is to characterize and functionalize pulse proteins isolates produced by salt extraction coupled with ultrafiltration/diafiltration and relate functional properties to texturization potential. Two functionalization techniques – cold plasma and transglutaminase – will be conducted to induce polymerization that could potentially enhance texturization. The second objective is to evaluate the impact of different extrusion parameters of twin-screw extruder on the physicochemical and mechanical properties of the texturized proteins. The last objective is to determine the sensory quality of the texturized proteins. This research is the first study to investigate the impact of mild isolation process and polymerization treatments on the texturization potential of pulse proteins.
Currently, structural and functional characterization are being conducted on all in-house and commercial protein ingredients. Results so far indicated that ChPI have higher level of legumin and potentially superior functional properties compared to PPI. (03/2021)
Legumes of the Future: Developing Methodologies and Germplasm to Enhance the Functionality and Nutritional Quality of Pea Protein
Funded by Foundation of Food Agricultural Research
Principal Investigators: Robert Stubar & B. Pam Ismail
Researcher: Sungil Ferreira
Plants are an important source of protein in the human diet, but most plant-derived proteins are not as nutritionally balanced and are not as readily digested as animal proteins. Dried yellow peas are a good source of protein with applications in many food products. Pea protein has lower allergenicity and fewer potential side effects than soy protein, the most common plant protein ingredient, but lags behind soy protein in terms of protein functionality and nutritional quality. This project focuses on the development of peas with improved protein profiles and higher productivity in diverse environments. The project will survey the genetic variation of public yellow pea accessions and develop improved and high-throughput methods for predicting pea seed protein functionality. We will also assess yellow pea accessions for protein digestibility and amino acid profile. Furthermore, we will identify regions of the pea genome that control storage protein content and distribution ratios and initiate breeding activities to select for improved pea protein functionality and digestibility. Products from this work will include new tools and knowledge to support the breeding of pea cultivars with these traits. These products will find immediate applications in a rapidly expanding plant-based food market. (03/2021)
Optimization of Extraction of Camelina (Camelina sativa) Protein and Screening for Varietal Differences
Funded by Forever Green Initiative
Principal Investigator: B. Pam Ismail
Researcher: Vaidehi Narkar
This research focuses on developing and optimizing industry-feasible extraction methodology to produce camelina protein isolates with optimal yield and purity as well as structural integrity. The extraction methods have been optimized to produce camelina protein isolates from Joelle line with a purity of 83-84% and a protein yield of 55-63%. The conditions for pH extraction were optimized to improve the color of the protein isolate without any protein polymerization. Currently, there are concerns about the functional properties of plant-based proteins. Addressing these concerns is important for the application of plant-based protein ingredients in food systems. Thus, this research project assesses the functional properties including solubility, water holding, gelation, and emulsification, as impacted by the structural characteristics of camelina protein. The camelina protein isolates from Joelle line demonstrated superior or comparable emulsification properties to commercial soy and pea protein isolates. Gelation properties of alkaline extracted camelina protein was acceptable and better than that of pea protein.
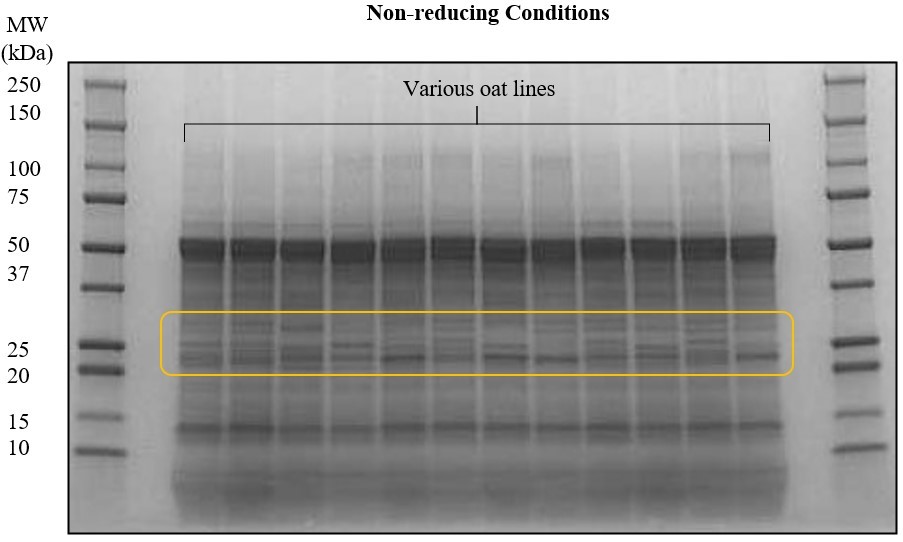
Diverse camelina lines are also being evaluated for protein functionality. Traditional plant breeding efforts at the University of Minnesota have identified camelina lines with potential differences in protein composition and profile. The protein profiles of 40 camelina lines, representing different growing conditions and origins, were screened using SDS-PAGE. Protein isolates produced from selected camelina lines will be used for further investigation of effect of breeding line on protein functionality. Structural and functional characterization of camelina lines will feed into the breeding program that aim at developing camelina protein for different food applications.
The development of quality-driven protein isolation protocols and optimal camelina lines for food use will advance its commercialization as an alternative and functional plant-based protein source. The knowledge and methodologies acquired through this research could also be applied to other protein sources and systems. (04/2021)
Impact of Selective Breeding, Extraction Conditions, and Modification Technologies on Pennycress Protein Structural, Functional, and Nutritional Characteristics
Funded by Forever Green Initiative
Principal Investigator: B. Pam Ismail
Researcher: Rachel Mitacek
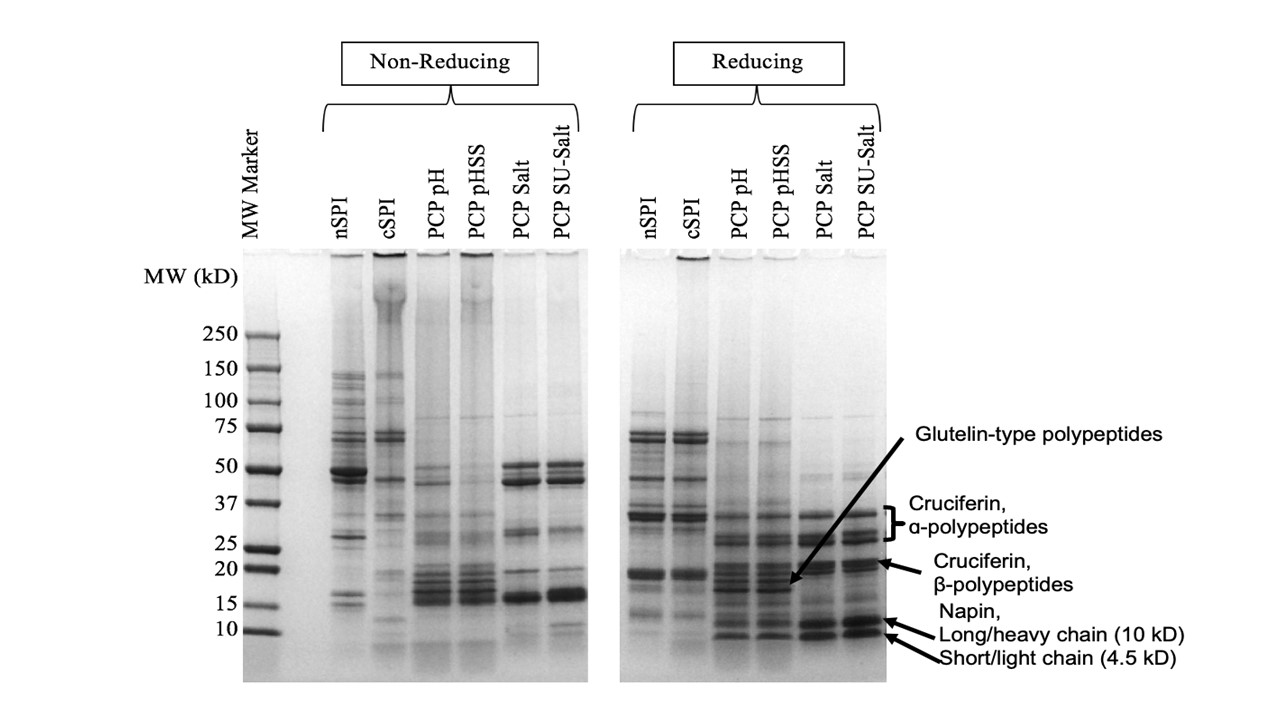
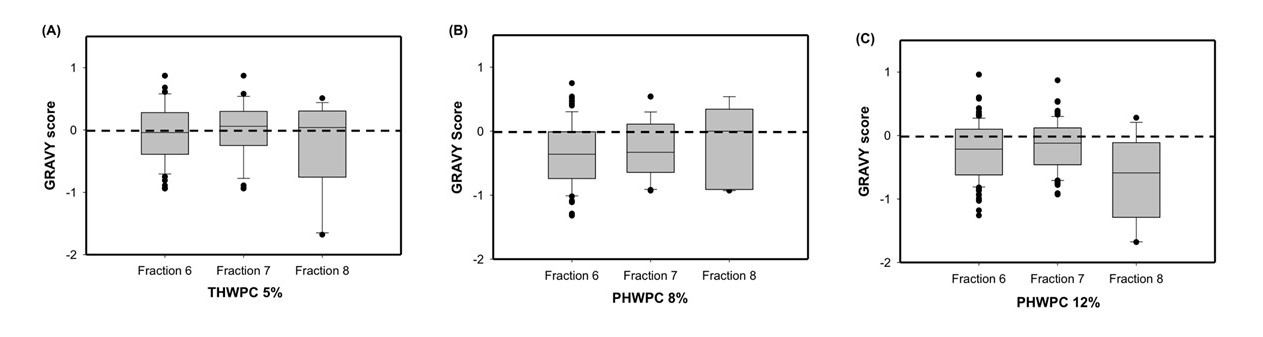
Pennycress (PC), a sustainable, short-season cover crop with vast environmental benefits, is a novel protein source. To develop a commercially-viable protein ingredient, effective protein extraction methods that preserve structural, functional, and nutritional attributes, as well as possible applications, need to be investigated. The objectives of this research were to 1) optimize protein extraction methods that maximize purity and yield, and characterize the influence of extraction method and scale-up on structural, functional, and nutritional attributes 2) evaluate the effect of traditional breeding on PC protein structure & functionality 3) investigate the enzymatic and atmospheric cold plasma (APC) effects on PC protein potential for texturization. Alkaline extracted protein was more denatured and polymerized, with higher surface hydrophobicity and aggregation, resulting in lower overall functionality compared to salt extracted protein. Scaled up, salt extracted protein had comparable and sometimes better functionality compared to commercial soy protein isolate. Enhancing texturization potential is currently underway. (03/2021)
nSPI: native soy protein isolate
cSPI: commercial soy protein isolate
PCP pH: pH extracted pennycress protein isolate
PCP pHSS: pH extracted with 0.1% sodium sulfite pennycress protein isolate
PCP Salt: Salt extracted pennycress protein isolate
PCP SU-Salt: Scaled-up salt extracted pennycress protein isolate
Protein profiling and characterization of diverse oat lines
Funded by PepsiCo
Principal Investigator: B. Pam Ismail
Researchers: Sungil Ferreira & Nigel Kang
Oat (Avena sativa) is the seventh most abundant cereal produced globally; however, it accounts for only under 1% of total cereal production. In recent years, the demand for sustainable, protein-rich, and nutrient-dense foods has increased due to shifts in consumer interests. Oats are notable among cereal grains for their superior nutritional value and well-balanced amino acid composition. Oats separate themselves from other cereals by having lower allergenicity and a higher content of protein (15-20%), unsaturated fatty acids, soluble fiber (beta-glucans), and antioxidants. For these reasons, there are opportunities for oat research and production to grow, especially for human food applications. Therefore, the objective of this study is to evaluate a set of genetically diverse oat samples and correlate protein profiles with environment and genetic variation parameters. About 200 oat lines will be screened to characterize structural differences by proximate analysis, Osborne fractionation, SDS-PAGE, size-exclusion HPLC, and LC-MS/MS. In particular, the percent distribution of protein fractions in oats—namely, albumin, globulin, prolamin, and glutelin—will be contrasted and studied closely. Following the screening, a subset of oat lines will be further characterized for protein quality by in vitro assays and functionality by evaluating solubility, gelation, emulsification, and foaming properties. Variability within lines will be assessed between environment and growing location by genetic marker-based clusters. This research will be used to ultimately cross-breed oat varieties with enhanced protein nutritional quality and functionality. (03/2021)
Extraction and Modification of Pea Protein for Improved Functionality, Flavor, and Nutrition
Funded by USDA ARS
Principal Investigators: B. Pam Ismail, Gary Reineccius, & Daniel Gallaner
Researchers: Lucy Hansen, Yara Benavides-Paz, & Joseph Eggers
Extraction conditions including solubilization pH, isoelectric precipitation pH, solubilization duration, number of solubilizations, and use of dialysis were optimized for pea protein isolate (PPI) production by alkaline solubilization coupled with isoelectric precipitation (pH-extraction), based on protein purity and yield as well as industrial feasibility. Similarly, purification conditions including use of ultrafiltration (UF) and dialysis, individually or combined, were optimized for PPI production by salt solubilization coupled with membrane filtration (salt-extraction). Optimized benchtop methods were then scaled-up to pilot plant production. The structural characteristics and functional properties of the scaled up PPIs were compared to commercial PPI. Results demonstrated successful optimization and scalability of two pea protein extraction methods: alkaline solubilization with isoelectric precipitation and salt solubilization coupled with membrane filtration. Both methods achieved high protein purity and yield, and superior functionality to commercial isolates. Nutritional evaluation of the isolates is underway. Samples during pilot plant production were collected for flavor analysis as described below.
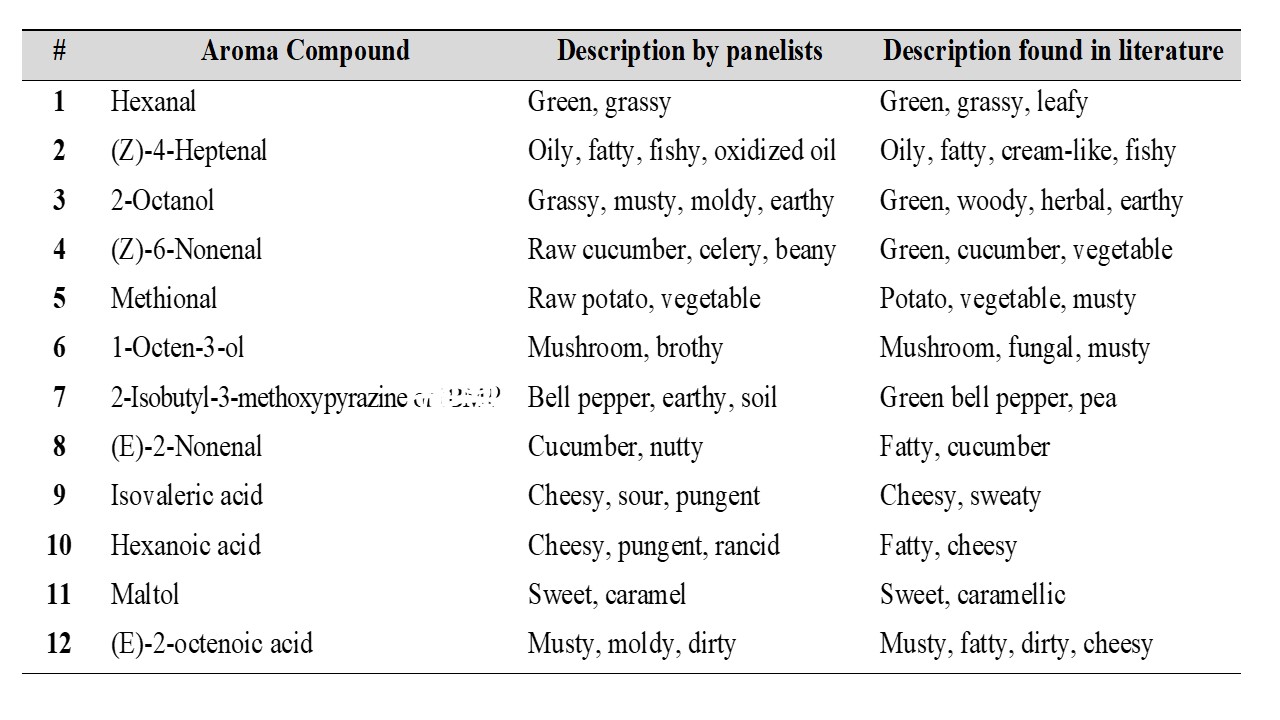
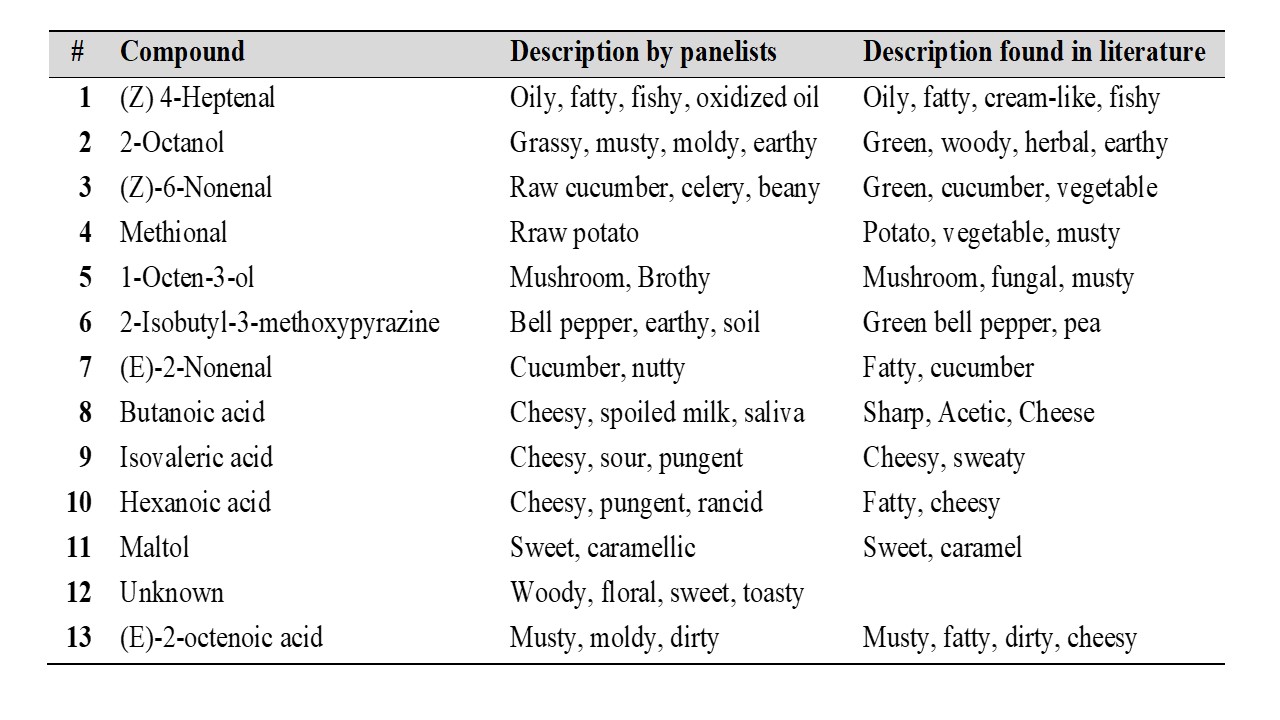
Three different methods to isolate aroma compounds from pea flour were tested: SBSE (Stir Bar Sorptive Extraction), SAFE (Solvent Assisted Flavor Evaporation) and SPME (Solid Phase Microextraction). SAFE was found to provide a more complete aroma profile of pea flour compared to SBSE and SPME. In order to monitor the aroma profile during the production of pea protein isolates by salt and pH extraction, samples were collected at several steps throughout the extraction process. The aroma compounds present in each sample were then isolated by using SAFE and analyzed by GC-MS-O (Gas Chromatography – Olfactometry). Three panelists were recruited and instructed to record the retention time, sensory descriptors and to rate the odor intensity of each odorant eluting from the sniffing port. The aroma compounds detected by at least two panelists with an odor intensity ≥ 3 (using a general magnitude scale where 0 corresponds to “no sensation” and 6 corresponds to “strongest imaginable sensation”) in at least one of the steps were then identified using retention indices, mass spectral data. The tables below show the aroma compounds that were detected and rated with a high odor intensity by the panelists (≥ 3) during the production of pea protein by salt and pH extraction.
We are currently working on analyzing the effect of processing conditions on the relative amount of the aroma compounds present in the samples. Preliminary results showed that there are not compounds that suddenly appear or disappear along the process and therefore, the processing steps do not completely remove or promote the formation of aroma compounds. (03/2021)
Protein-Flavor Interactions in Food Matrices
Funded by Several External Sources
Principal Investigator: Gary Reineccius
Researchers: Vaidhy Anantharamkrishnan & Igor Shepelevs
Adding flavorings to high protein foods and beverages leads to interactions, generally reducing overall product acceptance and shelf life. Therefore, it is pertinent to understand protein-flavor interactions to formulate consumer acceptable products. There has been a lot of research studying these multifaceted interactions, particularly on the “temporary” protein-flavor interactions (hydrophobic, hydrophilic, and ionic), but very little research has been done on a more permanent interaction: covalent bonding. Studying the covalent linkages is important because they are irreversible, and therefore will alter the flavor profile in an irreversible manner. This research aims to determine the covalent bonds that are formed between the side chains and terminal amino acids of plant proteins (soy, pea, canola) and aroma compounds. The extent and rate of these chemical reactions are monitored by MALDI-TOF MS and UPLC/ESI/TOF-MS. This study will provide understanding of the broad range of interactions of different flavor molecules with proteins under different conditions, as the type and rate of interaction are expected to vary with functional groups, flavor compound usage levels, protein structure, amino acid composition, pH, water activity, storage temperature, and food composition
Functionalization of Plant Proteins
Funded by Several External Sources
Principal Investigator: B. Pam Ismail
Plant protein (other than soy) functionality may not outperform commonly used protein ingredients. Accordingly, we are investigating ways to enhance functionality. Functionalization approaches of various plant proteins currently investigated include targeted and limited enzymatic modification utilizing various common and unique enzymes, Maillard-induced glycation, and chromatographic fractionation. We will also be investigating physical means such as the use of cold plasma. Functionalization is assessed via various analytical tools including spectroscopy, microscopy, proteomics, and chemical assays to evaluate the impact of functionalization on molecular changes. Structural/functional relationships are assessed to achieve desired modification for targeted and unique applications.
Completed Protein Research Projects
Enzymatic modification of pea protein isolate for improved functionality
Funded by USDA ARS
Principal Investigator: B. Pam Ismail
Researcher: Maneka Malalgoda
In recent years, the demand for plant protein ingredients has increased rapidly. Although ingredients such as soy protein isolate are widely used as a food ingredient, novel ingredients such as pea protein isolate are gaining interest. However, in comparison to soy protein isolate, pea protein isolate is not as functional. In this study, seven enzymes were investigated under optimized conditions for hydrolysis, to produce hydrolysates with degree of hydrolysis between 5-7%, and of these, two enzymes were used in a sequential hydrolysis. In general, upon hydrolysis smaller proteins and peptides were produced, which displayed lower charge and higher surface hydrophobicity. In terms of functional properties, hydrolysis did not improve gelation, and solubility was generally lower in the hydrolysates, with the exception of Trypsin-flavorzyme hydrolysate at pH 3.4 under non-heated conditions. However, emulsification properties were improved as a result of hydrolysis. This study shows the versatility of enzymes in the context of improving functionality of pea protein isolate. Additionally, the results show that targeted improvement of functionality can be achieved through the use of specific enzymes under optimum conditions for partial hydrolysis. For example, the Trypsin-flavorzyme showed improved EC, EAI and ES as well as FC and FC. Overall, this work emphasizes the need to investigate novel enzymes as well as hydrolysis with multiple enzymes as a potential way of improving pea protein functionality in food systems. (03/2021)
Improving the Functionality of Low Protein Streams for High Value Applications
Funded by Midwest Dairy Association
Principal Investigator: B. Pam Ismail
Researcher: Chelsey Hinnenkamp
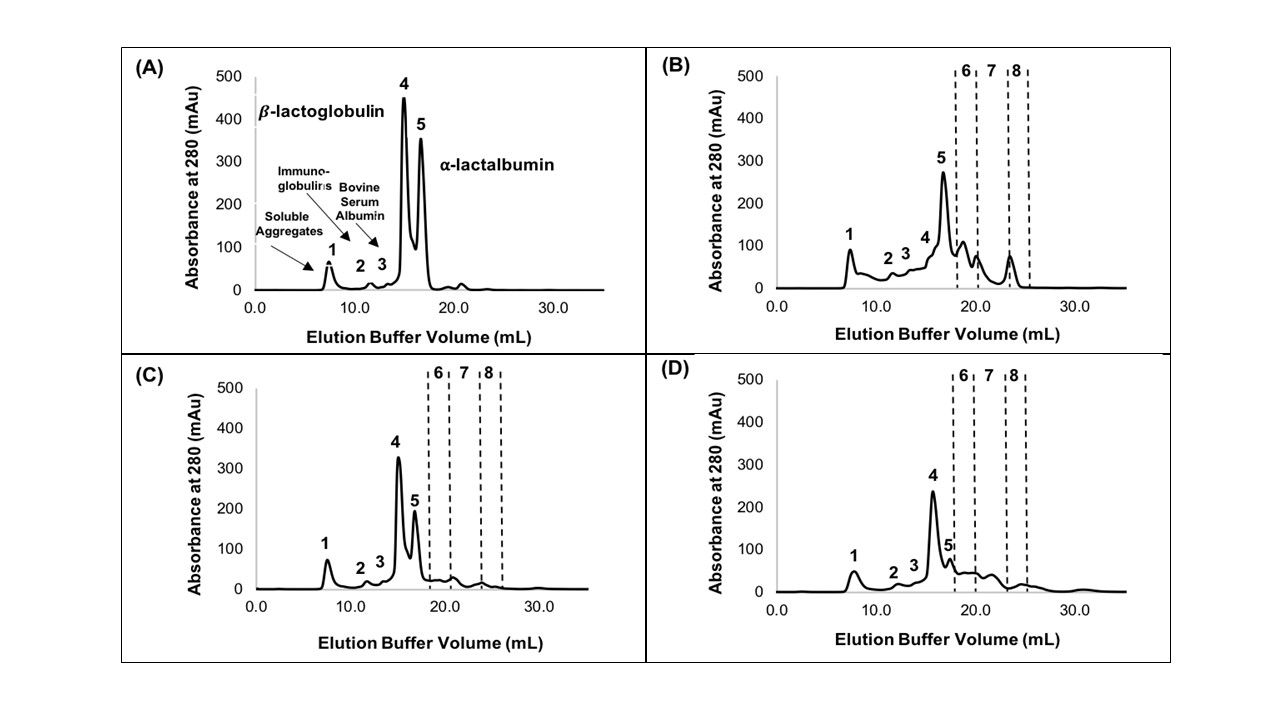
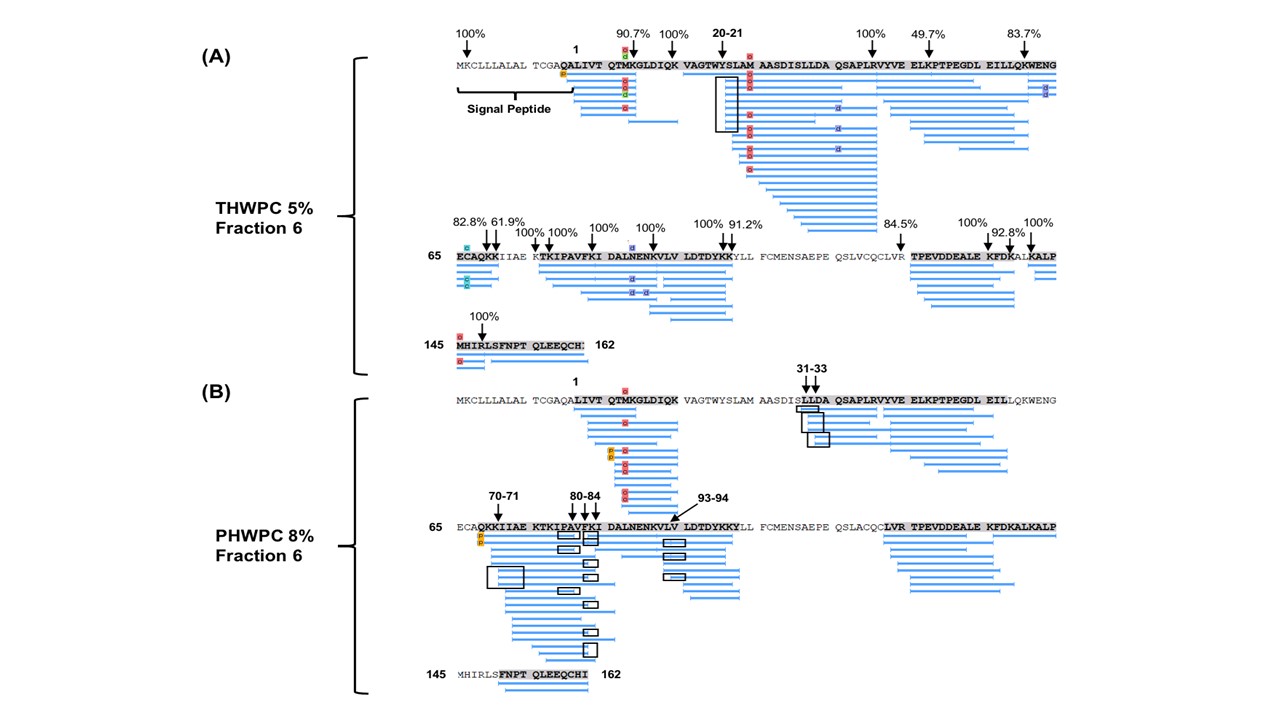
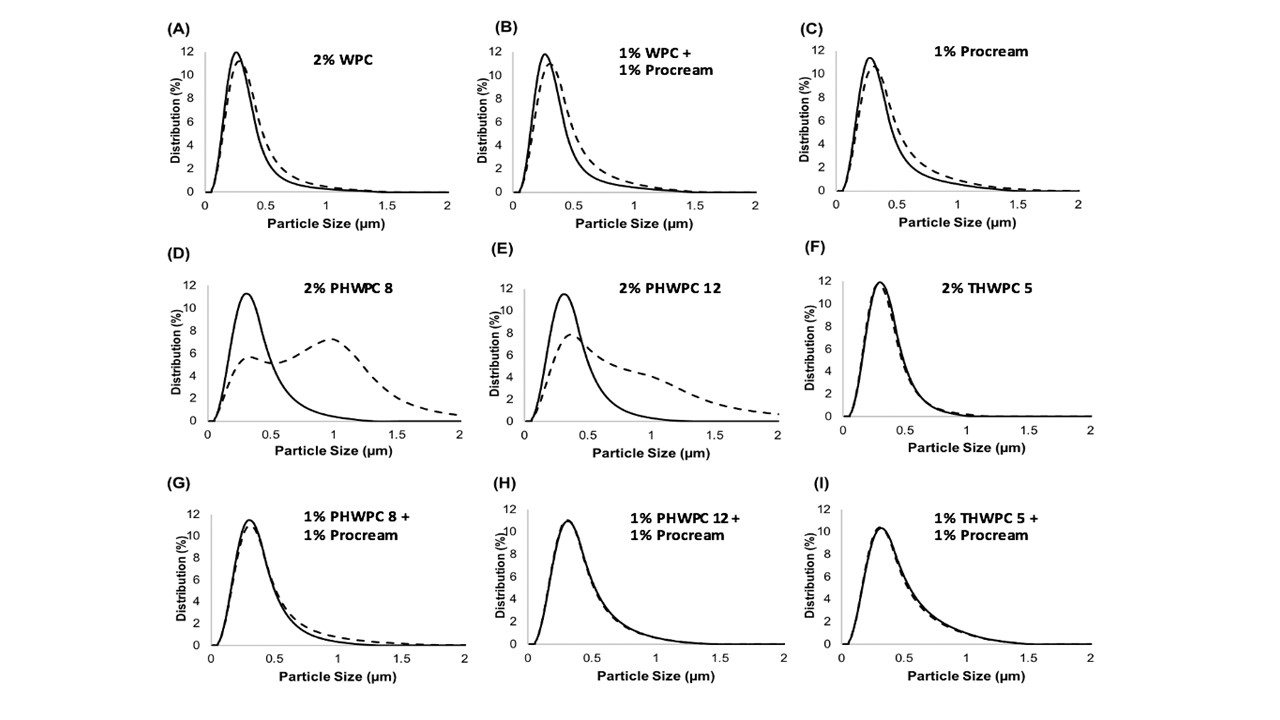
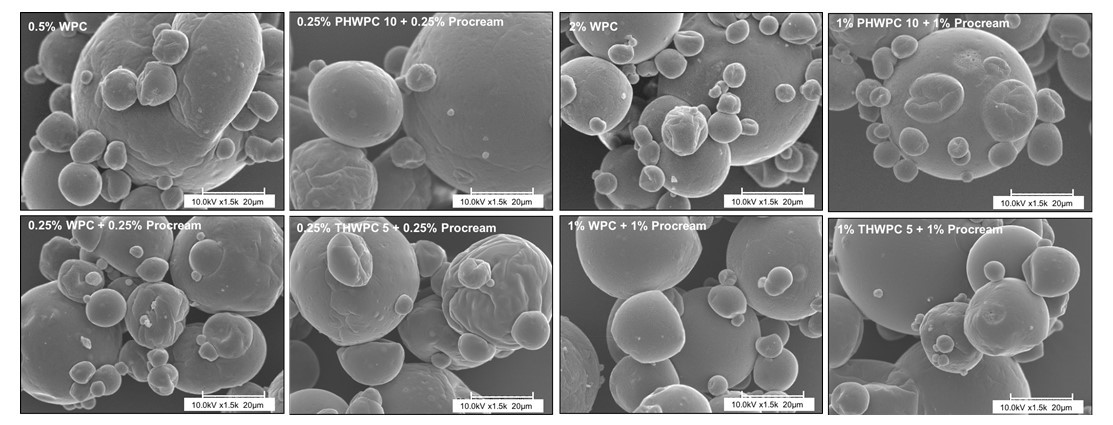
The industry solution to the increased consumer appetite for protein and protein enriched products has been to utilize protein concentrates (> 80% protein) and isolates (>90% protein) in their formulations to enhance nutrition, functionality, and overall protein content. However, the production of these high protein ingredients generates coproducts, such as Procream, a phospholipid-rich whey coproduct, that are underutilized due to lower protein content and functionality. A potential valorizing opportunity for Procream is to blend it with functionally enhanced WPC for high value applications like microencapsulation of oils rich in omega 3 fatty acids, such as fish oil. By leveraging inherent characteristics, blending Procream with hydrolyzed whey protein has the potential to improve interfacial activity and antioxidant activity of whey protein, which have an augmentative effect on microcapsule stability and overall success. There exists a need to not only understand the performance of these blended systems in microcapsule systems but to understand the interactions between Procream and intact and hydrolyzed WPC driving that performance. This project employed a combination of proteomics and bioinformatics to better characterize protein hydrolysis and related protein structural characteristics with inherent properties of the blends to understand the impact of blending on emulsion and encapsulation systems.
Results demonstrated for the first time that blending of Procream with hydrolyzed WPC had a synergistic effect on emulsion stability, while Procream demonstrated potential as an acceptable replacement for more expensive whey protein ingredients in microcapsule systems. The “molecular to applied” approach employed allowed for better optimization of whey protein hydrolysis for emulsification and encapsulation application. This approach also provided an understanding on how interactions between Procream and intact or hydrolyzed WPC contributed to emulsion stability and microcapsule performance. The “molecular to applied” approach of this project can be expanded to valorize plant protein coproducts and to help create markets for new plant protein streams, while providing clean-label, protein-based solutions to food industry challenges. (03/2021)
Extraction, Modification, and Structural/Functional Characterization of Camelina Protein
Funded by GMI
Principal Investigator: B. Pam Ismail
Researcher: Claire Boyle
Camelina sativa, a sustainable, short-season cover crop high in protein (30%), is gaining interest due to the increasing global demand for protein-rich products and for novel plant proteins. In order to create a functional, market-viable ingredient, an efficient means of protein extraction needs to be established for camelina. The aim of this project was to evaluate two protein extraction approaches, alkaline and salt extraction, and determine their impact on the structural and functional properties of camelina protein. Protein yield, content, structural characteristics, and functional properties of the produced camelina protein concentrates (CPC, 70-80% protein) and hydrolysates (CPH) were assessed and compared to soy protein isolate (SPI). Compared to alkaline pH extraction, data showed that salt extraction produces less denatured and more functional CPC, composed mainly of cruciferin and napin proteins. The functionality of the salt extracted CPC was comparable and sometimes better than that of SPI. Overall, results revealed the potential of camelina as a novel source of functional plant protein that might gain a position in the protein market, and possibly compete with soy protein for several applications targeting the use of plant proteins. Further functionalization of camelina protein for various applications is underway.
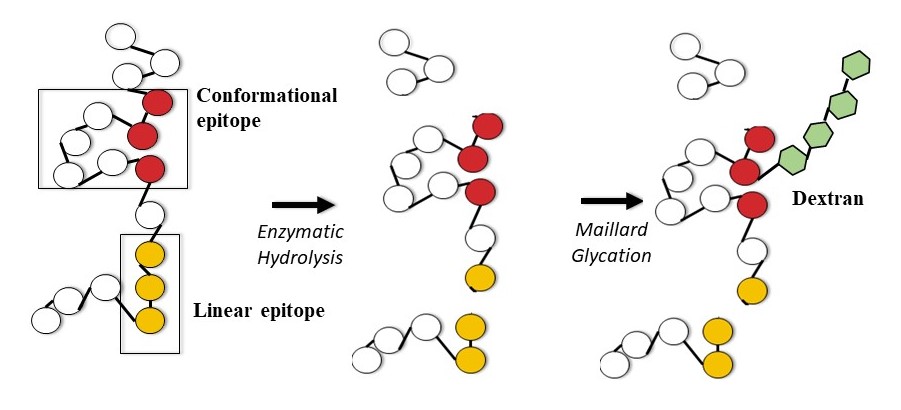
The Effect of Limited Enzymatic Hydrolysis and Maillard-Induced Glycation on Soy Protein Immunoreactivity and Functionality
Funded by Several External Sources
Principal Investigator: B. Pam Ismail
Researcher: Chelsey Hinnekamp
Soy protein is a functional protein of high nutritional quality, yet it is one of the “Big 8” food allergens. Accordingly, food manufacturers are seeking alternatives or solutions to the allergenicity of soy protein. To explore hypoallerginization of soy protein while maintaining functionality, this study evaluated the potential of combining two controlled protein modification approaches, limited/targeted enzymatic hydrolysis and partial Maillard-induced glycation. The impact of molecular/structural changes on immunoreactivity and functionality was assessed. Changes in immunoreactivity were assessed with immunoassays utilizing human sera. Limited enzymatic hydrolysis significantly reduced immunoreactivity, with reductions ranging from 20 – 75%, depending on the enzyme used and sera variability, while Maillard-induced glycation had a variable, at times negative, effect on immunoreactivity. To determine the effect of molecular changes upon modification on immunoreactivity, a proteomics approach was employed: two-dimensional gel electrophoresis and liquid chromatography-tandem mass spectrometry (LC-MS/MS) were coupled with two-dimensional immunoblotting. The modification did not cleave/mask all of the allergenic epitopes. The residual immunoreactivity was due to intact epitopes found on peptides cleaved from both β-conglycinin and glycinin subunits. Moreover, individual variability among sera was observed due to differential reactivity to various subunits. On the other hand, the targeted, dual modification had minimal effect on functionality of the protein. This study highlighted the importance of targeting individual variability in immunoreactivity and understanding the structural impact of modification when optimizing conditions to develop a functional, hypoallergenic soy protein ingredient. Additional work to achieve maximal reduction in immunoreactivity while maintaining functionality is under way.
Proteomics Approach to Characterize Intermediate Wheatgrass Proteins
Funded by The Forever Green Initiative
Principal Investigator: B. Pam Ismail
Researcher: Chathurada Gajadeera
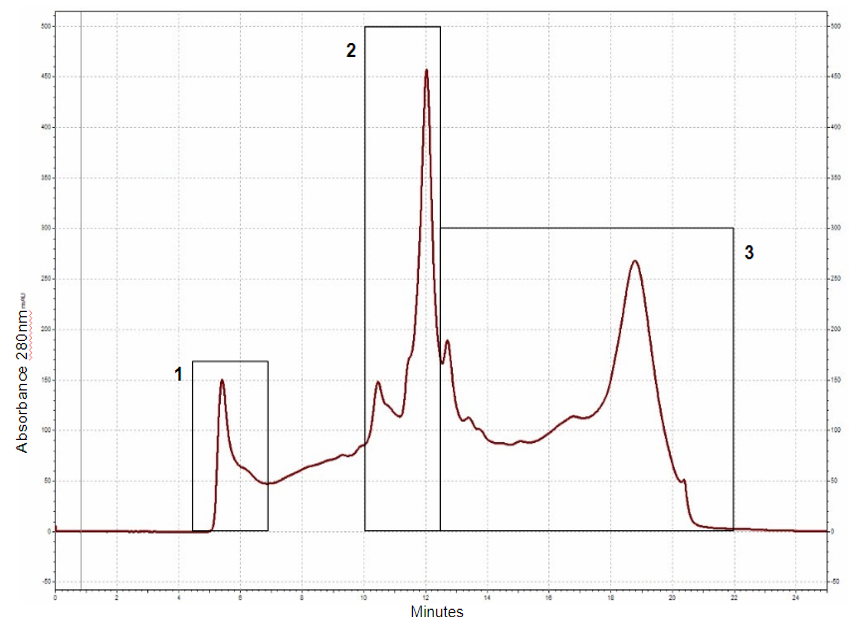
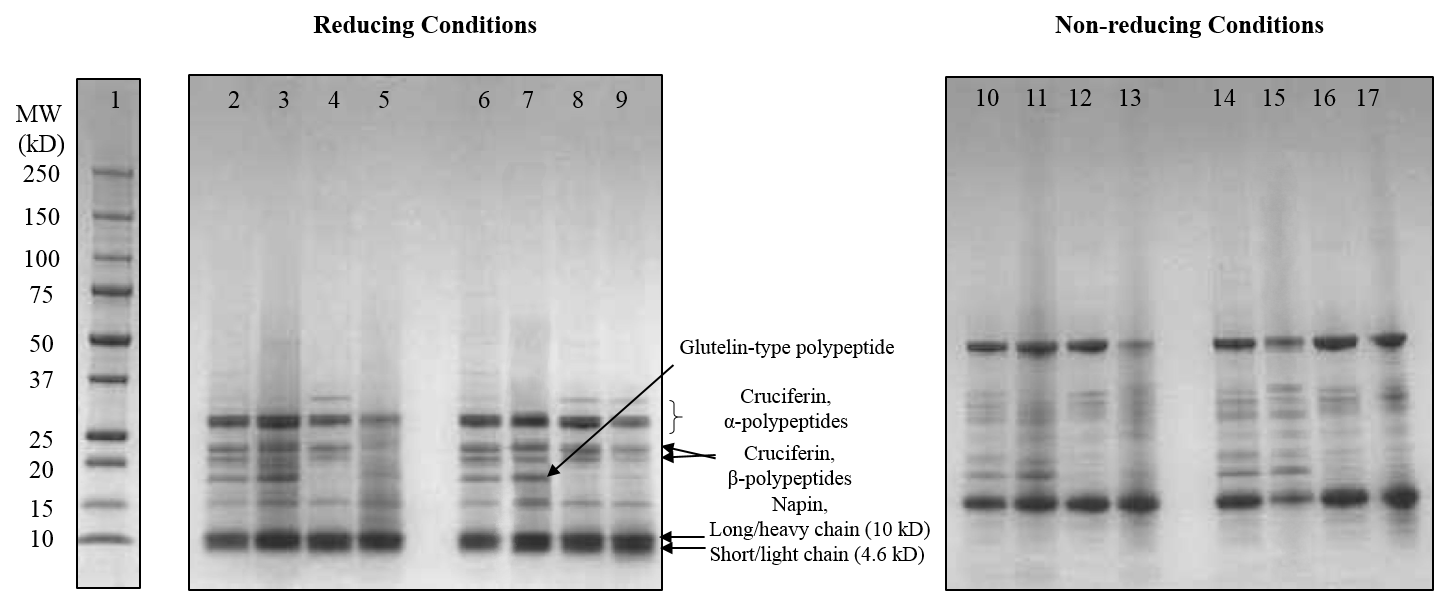
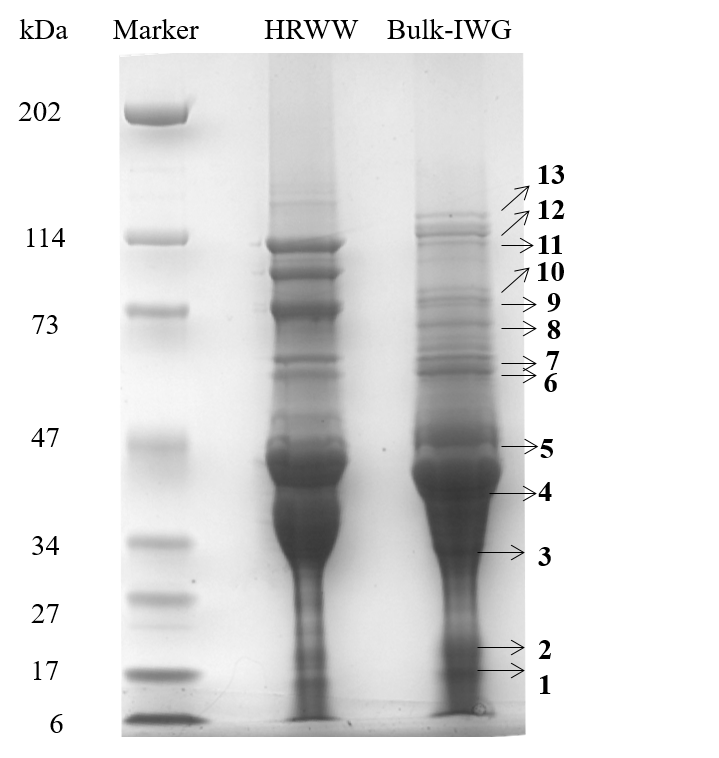
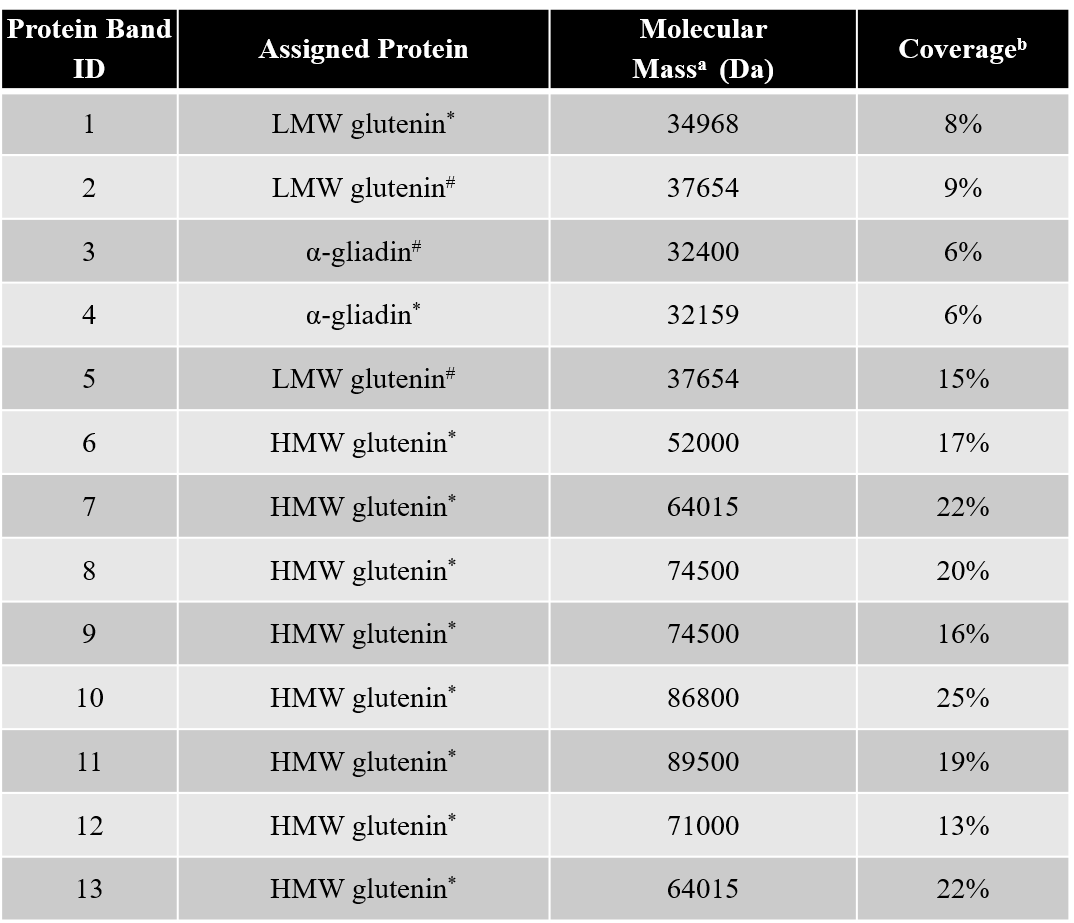
Thinopyrum intermedium, commonly known as intermediate wheatgrass (IWG), is a perennial crop with favorable agronomic characteristics and nutritional benefits. IWG is relatively high is protein content compared to wheat (17-21% compared 10-15% on dry basis), yet it is deficient in high molecular weight glutenins (HMWG), responsible for dough strength. The aim of this study is to characterize IWG proteins using a proteomics approach to elucidate impact on functionality and identify potential applications. Molecular weight distribution of IWG proteins are analyzed using size exclusion high performance chromatography. The figure below shows SDS-PAGE visualization of gluten proteins selected for chymotrypsin in-gel digestion for subsequent molecular identification by LC-MS. Isolated protein fractions are analyzed and identified by gel electrophoresis and LC-MS/MS respectively. Preliminary results showed that, in contrast to wheat, IWG had lower percentage of SDS-unextractable high molecular weight polymeric proteins (uHMWPP). Relative quantity of soluble albumins and globulins were higher in IWG samples than that of wheat. HMWG, low molecular weight glutenin (LMWG), α, β, γ and ω–gliadins, and albumins and globulins were identified by LC/MS-MS as major proteins in IWG. These proteins were considerably different in their molecular weights in contrast to those of wheat controls. Similarly, the protein distribution and molecular weights of HMWG subunits in the IWG bulk sample were considerably different from wheat. HMWG present in IWG were of lower molecular weight, approximately 60 kDa compared to 80-120 kDa in wheat. The lower uHMWPP content in IWG, suggest that it might not be suitable for applications that require dough rising properties. The detailed protein characterization of IWG, will be used to identify other unique applications to expand its market potential.
Identification of key volatile compounds contributing to pennycress aroma
Funded by Several Funding Sources
Principal Investigator: Gary Reineccius
Researcher: Peishan Luo
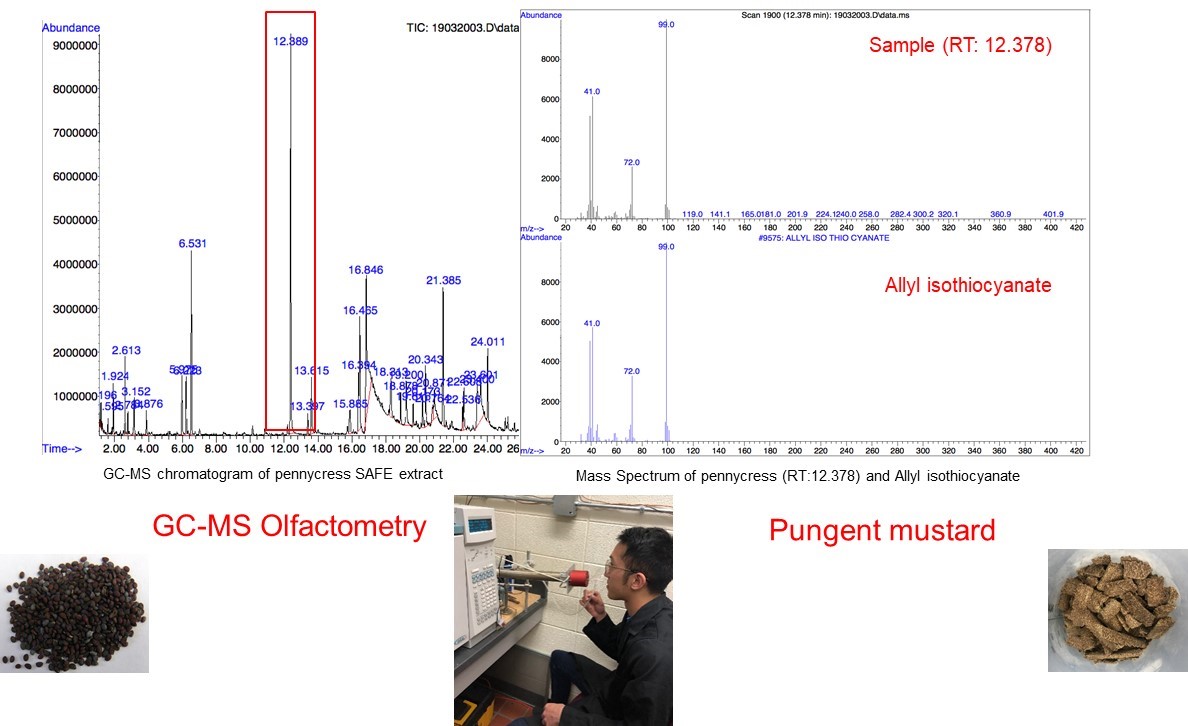
As a member of the mustard family (Brassicaceae), pennycress has an unpleasant “mustard-like” aroma, which is hard to be accepted by consumers, largely limiting its application in food industry. However, no published literatures has reported the volatiles analysis of pennycress. This research aims to identify key volatile compounds that contribute to pennycress aroma, thus making it more possible to design breeding programs or processes to minimize the undesirable aroma. To extract volatile compounds present in wild type pennycress seeds, solvent extraction and solvent assisted flavor evaporation (SAFE) are used, and subsequently, volatiles are detected and analyzed by gas chromatography-olfactometry (GC-O), coupled with aroma extraction dilution analysis (AEDA). Consequently, key volatile compounds are identified using retention indices, aroma descriptors, and standard chemicals verification. Identification of these volatiles may lead to determining the metabolic pathways of their production in the seed and changes in flavor development during protein isolation.